We can explore associations between genes and phenotypes
Genomic information plus clinical information in a large database allows us to identify genes associated with disease variants and truly begin precision medicine. In pharmacoepidemiology this will mean that we can identify patients who benefit most or are at greatest risk of an adverse event from a specific treatment.
The power of linking genomic information with clinical information is demonstrated in a recent study by Karnes et al 2017. They examined variations in major histocompatibility complex (MHC) and human leukocyte antigen (HLA) genes by linking information from DNA biobanks databases with EHR records for almost 30,000 people. Most importantly, they discovered previously unidentified associations between gene variation and disease severity for several conditions. They identified associations with type 1 diabetes and ankylosing spondylitis, among other conditions. Most importantly, they discovered several previously unidentified associations with MS and cervical cancer, as well as associations between gene variation and disease severity for several conditions.
The heatmap crosstabulates HLA alleles with a range of disease diagnoses with darker reds being stronger positive associations (the allele increases the probability of having the disease or phenotype) and darker blues being stronger negative associations (the allele is associated with a protective effect against the disease or phenotype).
The authors note that this is one of the first systematic studies of clinical phenotypes with HLA variation in a single population.
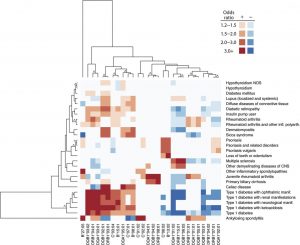
Heatmap of four-digit HLA allelic associations with P < 1 × 10?5.
Data are publicly available www.phewascatalog.org